5JMU
 
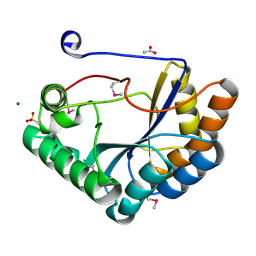 | | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 | | Descriptor: | ACETATE ION, MAGNESIUM ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Tan, K, Gu, M, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-29 | | Release date: | 2016-06-29 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The crystal structure of the catalytic domain of peptidoglycan N-acetylglucosamine deacetylase from Eubacterium rectale ATCC 33656 (CASP target)
To Be Published
|
|
4DYU
 
 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
4EAE
 
 | | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e | | Descriptor: | D-MALATE, Lmo1068 protein, SODIUM ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of a functionally unknown protein from Listeria monocytogenes EGD-e
To be Published
|
|
4DQ0
 
 | |
4EDH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg.
To be Published
|
|
6V6N
 
 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | Descriptor: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
4DZR
 
 | | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
3FBY
 
 | |
3G74
 
 | | Crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560 | | Descriptor: | Protein of unknown function, SULFATE ION | | Authors: | Tan, K, Sather, A, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560
To be Published
|
|
3FQ6
 
 | |
3FZ4
 
 | | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Putative arsenate reductase, ... | | Authors: | Tan, K, Hatzos, C, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The crystal structure of a possible arsenate reductase from Streptococcus mutans UA159.
To be Published
|
|
5CJJ
 
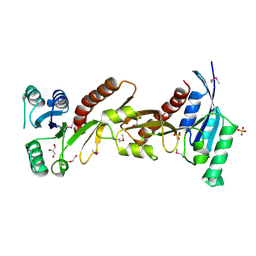 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-14 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
5CD2
 
 | | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114 | | Descriptor: | CHLORIDE ION, Endo-1,4-D-glucanase, GLYCEROL, ... | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114
To Be Published
|
|
3NZE
 
 | | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1. | | Descriptor: | CALCIUM ION, Putative transcriptional regulator, sugar-binding family | | Authors: | Tan, K, Zhang, R, Bigelow, L, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The crystal structure of a domain of a possible sugar-binding transcriptional regulator from Arthrobacter aurescens TC1.
To be Published
|
|
3OM8
 
 | | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable hydrolase | | Authors: | Tan, K, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas aeruginosa PA01
To be Published
|
|
3OCM
 
 | | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative membrane protein, ... | | Authors: | Tan, K, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The crystal structure of a domain from a possible membrane protein of Bordetella parapertussis
To be Published
|
|
3OCO
 
 | |
3OBB
 
 | | Crystal structure of a possible 3-hydroxyisobutyrate Dehydrogenase from pseudomonas aeruginosa pao1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Yakunin, A.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3OD1
 
 | | The crystal structure of an ATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C | | Descriptor: | ATP phosphoribosyltransferase regulatory subunit, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER | | Authors: | Tan, K, Bigelow, L, Hamilton, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-08-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of aATP phosphoribosyltransferase regulatory subunit/histidyl-tRNA synthetase from Bacillus halodurans C
To be Published
|
|
3OLQ
 
 | | The crystal structure of a universal stress protein E from Proteus mirabilis HI4320 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | The crystal structure of a universal stress protein E from
Proteus mirabilis HI4320
To be Published
|
|
4NAS
 
 | | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446.
To be Published
|
|
4R7Q
 
 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4RN7
 
 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
4S1N
 
 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4
To be Published
|
|
4RNL
 
 | | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus | | Descriptor: | GLYCEROL, PHOSPHATE ION, possible galactose mutarotase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible galactose mutarotase from Streptomyces platensis subsp. rosaceus
To be Published
|
|
